Systems Biology & Biomedical Data Science Laboratory
We develop and apply data science approaches to understand health and disease for HUMAN | DATA | SOCIETY.
Research
Utilizing data for human society.
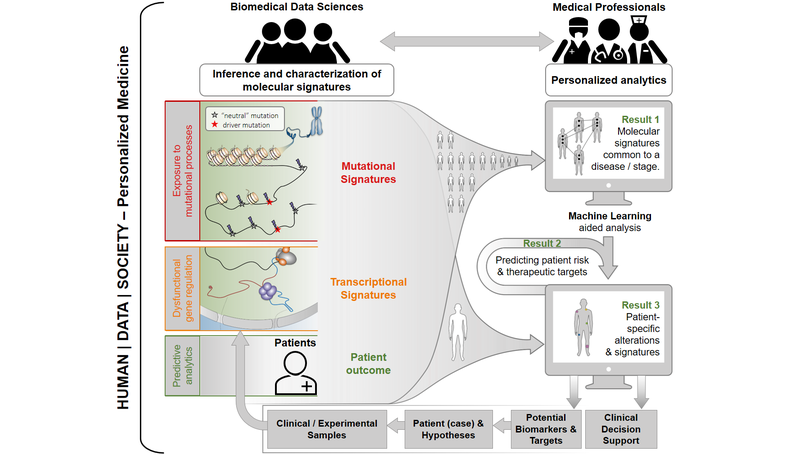
Personalized medicine can improve the lives of patients and clinical professionals lastingly. Thus, developing data science approaches, tools and systems for personalized analytics is one of the aims of our research group, which is a member of the recently established Centre for Human | Data | Society.
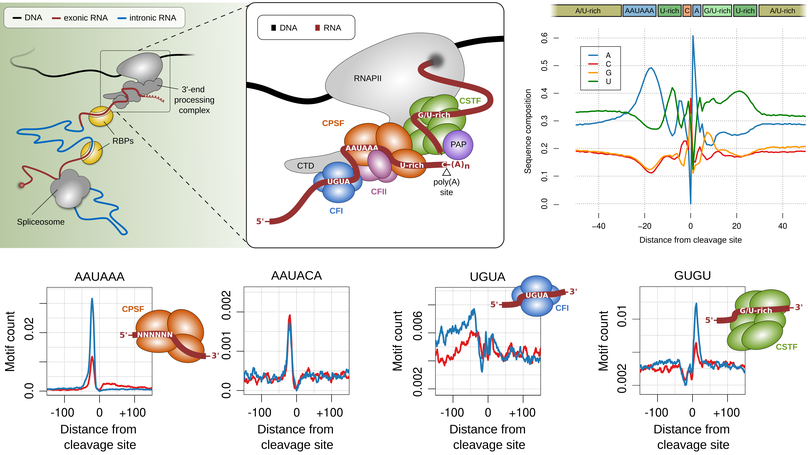
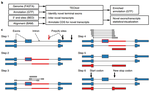
The 3' end of RNA polymerase II transcripts is generated by endonucleolytic cleavage and polyadenylation at 3' end processing sites, also termed poly(A) sites. The processing of 3' end processing sites is mediated by the so called 3' end processing complex, which is a huge machinery that consists out of several subcomplexes (CFI, CPSF, CSTF, CFII) that bind to specific sequence motifs in vicinity to poly(A) sites, the most prominent of which is the so called canonical poly(A) signal (‘AAUAAA’).
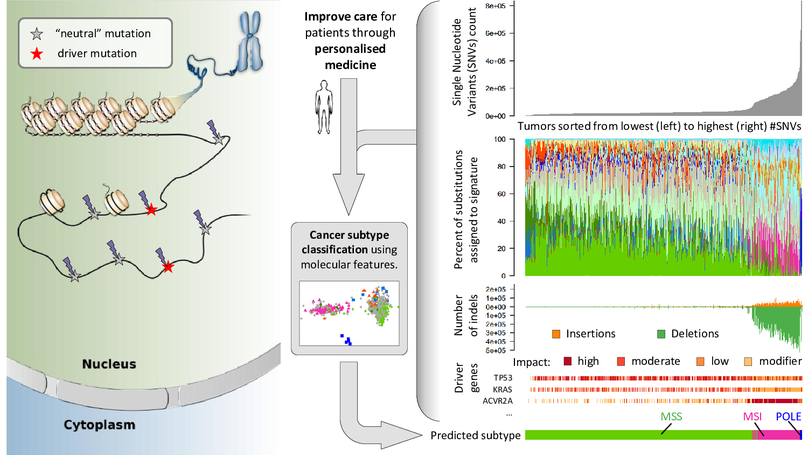
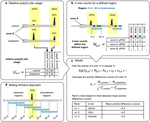
Tumors harbour various molecular features, such as tumor mutation burden (single nucleotide variants and indels), specific driver gene mutations and mutational signatures. We are subtyping cancers based on various molecular alterations at a fine level of detail and investigate their association with clinical outcomes.